Review Article - Oxidants and Antioxidants in Medical Science (2021)
The Influence of Various Forms of Selenium on Redox Processes, Gene Expression of Selenoproteins, Antioxidant Status in Biological Objects
Bityutskyy VS*, Oleshko OA, Tsekhmistrenko SI, Melnichenko OM, Tsekhmistrenko OS, Melnichenko YuO, Heiko LM, Oleshko MO, Kharchyshyn VM, Fedorchenko MM, Vered PI and Shulko OPBityutskyy VS, Department of Life Sciences, Bila Tserkva National Agrarian University, Ukraine, Email: voseb@ukr.net
Received: 05-Jul-2021 Published: 26-Jul-2021
Abstract
In this review, we consider literature data that offer the concept of the presence of a sensory physicochemical barrier between an organism with its functional genome and the environment (exposure), called the redox interface. Oxidation reduction of proteins (proteome) is considered as an important element of the organismâ??s adaptation to the environment. The positioning of Se and selenoproteins at the redox border offers an analogue to consider the more general interaction of the genome with exposomes. Data are presented showing the role of selenoproteins in various metabolic processes, such as redox dependent signaling. The collective functioning of these signaling connections in the body creates an adaptive network through which the genome uses resources and protects cells from environmental threats. The influence of neo organic and organic forms of selenium on the expression of selenoprotein genes, productivity, metabolic and immune processes in the body of aquaculture objects is described.
Introduction
In biological systems, the most common reactions are Redox Reaction (RR). The main feature of biological systems is that RR in most cases catalyzes proteins, which indicates the presence of genetic control over redox processes. Oxidoreductases, which catalyze the reactions of oxyreduction, have characteristic properties. The primary amino acid sequence of their apoenzyme determines the conformation of “pockets” specific for coenzymes. This interaction is a prerequisite for the catalytic activity of oxidoreductases. In the spatial organization of the protein, the interaction of amino acid residues (Cys, His, etc.) is of great importance, what determines the specificity and effectiveness of the intra and intermolecular electron transfer pathways.
The balance of redox processes (redox status) determines cellular redox homeostasis, on which bioenergetic and essential cell functions, including differentiation of proliferation, proteostasis, apoptosis, and autophagy, depend to a large extent [1]. The leading role in maintaining redox homeostasis is played by the ratio of the processes of generation and catabolism of reactive oxygen species (ROS), catalyzed by enzymes and enzymatic systems.
Their imbalance can lead to an increase in the level of intracellular ROS and an increase in oxidative processes, and ultimately to oxidative stress, which disrupts the harmonious cell defense system, which leads to instability of the genome and the onset of cancer [2] and other pathologies [3].
The most studied are redox systems in which the Cys HS groups are components of proteins, coenzymes, and also NADP H and perform the functions of the main redox elements. In this regard, the following formulations of the definition of redox systems are found. These are systems in which: a number of electron donors restore redox pairs of NAD and NADP; the terminal electron acceptor is O2, which is the source of H2O2 and other ROS; intermediate redox modules (Cys, Met), due to oxidation reduction, ensure the regulation of cellular functions [4,5].
Protein oxidation reduction (proteome) is considered as an important element of the organism’s adaptation to the environment [6,7]. By changing the redox state of the proteins, the action of environmental factors (reception) is perceived and the signal is transmitted to the genome (signaling). In this regard, the link between the exposome and the genome is a change in the redox state of the proteome. The redox state of proteins is modulated by a variety of oxidizing agents and reducing agents. The main oxidizing agent of proteins in aerobic organisms is oxygen. The active participation of oxygen in RR was made possible thanks to the extremely reactive ability of ROS, which are formed in a variety of biochemical processes both spontaneously and purposefully [8-10].
A number of ROS, including superoxide (O2•–), hydrogen peroxide (H2O2), peroxynitrite (OONO–) and hydroxyl radical (HO•), are all formed in the biological systems O2•– and H2O2 are produced enzymatically and participate as in reversible physiological transfer processes signals and in pathologies associated with oxidative stress [11]. Other ROS, such as OONO– and HO• are not considered signaling molecules because of their high reactive nature and irreversible modifications, but nevertheless can contribute to oxidative stress and pathological tissue damage [12].
Selenium (Se) is a redox metalloid that takes part in redox processes in the body. Selenium in compounds has different oxidation states (-2, +2, +4 and +6), which allows it to exhibit specific biological properties in systems with wide integrative functions. Thus, according to modern ideas, selenium, taking into accounts the new theory of omix [13], is a component of the redox interface through which the body interacts with environmental signals (exposomes) and reacts in accordance with them, supporting homeostasis at the level of epigenome, genome, metabolome and exposure. The concept of a redox interface was introduced to denote the many and often overlapping functional systems that serve to protect the reduced intracellular redox elements from oxidative environmental conditions [14].
DNA methylation is a common epigenetic mechanism by which gene expression is regulated. Selenium in the form of SeMet increases liver DNA methylation, possibly due to exposure to a single carbon exchange (methyl donor for DNA methylation), including a higher SAM/SAH ratio and mRNA level of serine hydroxymethyl transferase [15].
Se affects the genome by maintaining the stability of the genome. The complexity of the processes and forms of Se often interferes with an accurate understanding of mechanistic details. Using integrated network approaches can improve this understanding.
Se promotes the regulation of the functional genome through the activation and repression of transcription factors that control gene expression. Se in optimal concentrations blocks the activation of the transcription factor NF-κB, which regulates the expression of inflammatory genes [16,17].
Quantitatively, the most important relationship between the proteome and animal metabolome occurs through two key Se-containing amino acids, selenocysteine (SeCys, Seс) and selenomethionine (SeMet) [18]. Selenocysteine has a high biological activity and differs from Cys in that it contains Se in the form of selenol; at physiological pH, the selenol group is largely ionized to selenolate, which makes it a more reactive nucleophile than Cys thiol. Access to the large scale omics datasets (genomics, transcriptomics, proteomics, metabolomics, metagenomics, phenomics, etc.) revolutionized biology and led to systemic approaches that foster a deeper understanding of biological processes. In fact, redox integrative systems are defined as components of the physicochemical, enzymatic, and chelate barrier systems that protect the redox elements of a living organism from external influences.
Selenoproteome and Selenoproteins
Selenium (Se) is presented in two forms in eukaryotic proteins as the rare amino acid selenocysteine (SeCys) and selenomethionine (SeMet). The term selenoprotein is used exclusively for proteins containing SeCys residues, since this is the main biologically active form of selenium in proteins. SeCys is encoded by a triplet codon (UGA), which is usually interpreted as a termination codon during translation. The incorporation of SeCys into the polypeptide chain of proteins (like the 21st amino acid) is achieved using a special molecular insertion mechanism. The sequence of insertion of SeCys occurs with the participation of SeCys recognizing proteins as specific elongation factors and compounds with tRNA. SeCys is present as a stem loop in the 3´-untranslated region or UTR of the mRNA of selenoprotein [19].
The amount of selenoproteins (selenoproteomes) can vary in different species of living organisms [20]. It has been established that aquatic organisms usually have larger selenoproteomes than terrestrial ones, while mammalian selenoproteomes tend to reduce the use of selenoproteins.
Glutathione peroxidases (GPx) and thioredoxin reductases (TrxR) are the most studied selenoproteins [21]. They are an indispensable component of the cellular glutathione and thioredoxin systems and, therefore, important regulators of the intracellular redox environment [22,23], affect the balance of endocrine energy [24], affect insulin resistance [25]. Selenoprotein P (SelP) is the main protein in the body responsible for Se homeostasis and transport through the body [26]. Interestingly, SelP is present as two different isoforms in fish, SelPa and SelPb; they have noticeable differences at the structural level and probably play different roles in the fish homeostasis Se [27]. Depending on their biological function, selenoproteins are usually divided into two groups: irreplaceable and non-essential selenoproteins. The role of these proteins dictates a strict hierarchical regulation of their synthesis, which becomes evident in conditions of low Se availability, where the expression of non-essential selenoproteins is disrupted in favor of selenoproteins with important functions [28,29].
In mammals, 25 selenoproteins had been identified, in bony fish (Teleostei), up to 41 [30]. Some salmon species may have even more selenoproteins, due to the doubling of the entire genome that occurred during the evolution of this group [31].
Adaptation to Se variation requires a wide range of compensatory reactions. It has been established that only a very limited number of genes encodes Secontaining proteins (25 selenoproteins in humans, 24 in mice, but more than 40 in different fish species), therefore only a small amount of proteins has a direct effect.
Genes what encode selenoproteins are involved in various metabolic processes, such as redox dependent signaling. Selenium containing glutathione peroxidases, from the family of multiple isozymes, are encoded by several gene clusters (GPx1a, GPx1b, GPx4a and GPx4b) in bony fish [32], thioredoxin reductase isoforms (TRXRD1, TRXRD2) [33], protein folding and degradation (SEP15, SELS), metabolism (SEPP1, SPS1, SPCS), which, in turn, alter the regulation and expression of genes [34].
Although the incorporation of Se into these proteins is important for their functioning, it is also known that a deficiency or excess of Se regulates the transcription of these selenoproteins [35]. Selenoprotein P (SelP) is the main protein responsible for homeostasis and Se transport in body tissues [36]. It was established that SelP is present in the form of two different isoforms in fish (SelPa and SelPb). They have noticeable differences at the structural level and probably play different roles in the fish homeostasis Se [37].
Two new SelP genes, namely SelPa2 and SelPb2, were found in rainbow trout and Atlantic salmon. The multiple Se residues in SelP proteins (especially SelPa1 and SelPa2) are a unique characteristic of this selenoprotein and may indicate an increased need for selenium for this group of fish. Therefore, the optimal level of Se in the diet of salmon fish may require reevaluation.
The Effect of Various Doses and Forms of Selenium on the Expression of Selenoprotein Genes, Productivity, Metabolic and Immune Processes in Fish
The importance of selenium is evidenced by the fact that in 2017 the XI International Symposium on the Use of Selenium in Biology and Medicine (Stockholm, Sweden) was held in connection with the celebration of the 200th anniversary of the discovery of selenium by Berzelius. Currently, selenium and its various forms have not lost their significance, but are acquiring a new color due to advances in genomics, epigenomics, transcriptomics, proteomics and metabolomics.
The chemical form of the delivered Se can greatly affect its bioavailability and, therefore, the entire body. The main form of Se in the most common feed ingredients is SeMet, which makes up more than 50% of the total amount of Se in corn, soy, wheat, barley, etc [38]. It was established that inorganic forms of Se are less bioavailable than organic selenium compounds, and this means that they can be more easily excreted from the body. In addition, inorganic selenium compounds exhibit toxicity at lower concentrations compared to organic forms.
The main reaction that takes part in the enzymatic reaction of selenite is:
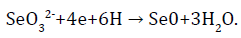
In this case, SeO32- ions serve as electron acceptors, and NADH dehydrogenase or components of electronic transport chains act as electron donors, restoring selenium to a zero-valence state [39].
The priority in feeding farm animals is the replacement of many trace elements which have been used for a long time in inorganic forms with organic analogues that are much more effective and biologically available [40]. Numerous scientific and industrial trials have shown that selenomethionine (an organic form of selenium) is an effective source of selenium to improve the health and productivity of animals and birds [41,42].
One of the most important forms of selenium for biological objects is selenocysteine (SeCys), the 21st amino acid in the genetic code [43,44]. The insertion of SeCys into a protein requires many additional factors, as well as the use of a specific stem loop sequence in mRNA to decode the UGA stop codon into a sense codon for SeCys [45-48].
The process of introducing SeCys into selenoproteins is bioenergetically expensive and SeCys facilitates the passage of this reaction, unlike cysteine (Cys) [49]. It had been found that SeCys, located at the C-terminus of the peptide, attacks the main chain carbonylamide much more easily than Cys, with the formation of selenoester [50]. The reason for the widespread use of SeCys is also that it can participate in reversible biological reactions. To date, discussions are underway on rationing selenium in compound feeds for fish. Many fundamental principles have been proposed for linking selenium concentrations in the whole fish organism or in the diet with adverse effects on fish. Different points of view of researchers form a differential approach to rationing the content of this element in the diet. Separate studies were examined and the basic principles for selenium concentrations in the whole body of the fish and in the diet, which were higher than those proposed by other researchers (4 μg/g in the whole body and 3-4 μg/g in the diet), were recommended. This article also recommends sharing the basic principles for cold water fish (6 μg/g for the whole body and 11 μg/g in the diet) and heat loving (9 μg/g for the whole body and 10 μg/g in the diet). Most selenium literature maintains a full body threshold of 4 μg/g in fish and 3 μg/g in the diet [51].
In Europe, the use of feed additives is regulated by European feed legislation. The maximum limit of total Se in animal feed, including fish feed, was set at 0.5 mg/kg (Council Directive 70/524/EC and amendments).
The European Food Safety Authority (EFSA) had published several scientific findings on the use of organic selenium yeast forms as feed additives. Based on the obvious higher bioavailability of organic Se compared to inorganic forms, it was found that the level of additives should be limited to a maximum of 0.2 mg/kg feed to ensure consumer safety. Subsequently, the European Union regulated the use of several Se feed additives, mainly selenized yeast, with an additive level of not more than 0.2 mg/kg feed [52,53].
The legislative distinction between general Se and its complementary organic forms requires the establishment of suitable methods for specifying Se. In addition, such methods will help to collect data on the presence of Se species in feed and fish, which is important for future risk assessments of feed additives [54].
A study on rainbow trout (Oncorhynchus mykiss) studied the uptake of the organic form of selenium as part of Sel-Plex and its effect on the expression of fish selenoproteomes [55]. Rainbow trout was fed a control diet containing Se at 0.9 mg/kg of feed, or the same diet enriched with three different Sel-Plex concentrations: 0.5 mg/kg, 4 and 8 mg/kg, which corresponds to the concentration of Se 1, 4 mg/kg (low-Se diet), 4.8 (middle class diet) and 8.9 mg/kg (high-Se) diet), respectively. The added additives of organic selenium (Sel-Plex) did not affect the survival and growth of fish. The distribution of selenium in organs is as follows: liver, kidney, muscle, and blood cells. With a high level of Se in the diet (4.8 and 8.9 g/kg) and a longer exposure time, the liver is not able to regulate the content of selenium, and the concentration of the element in the tissues increases. In order to investigate the body’s biological response to Sel-Plex supplementation, the authors studied the effect of Sel-Plex supplementation on mRNA expression of selected trout selenoproteins. The liver was the most sensitive tissue at the transcriptome level, followed by kidneys, blood cells, and muscles. This directly reflects the accumulation of Se in these tissues described previously, and additionally confirms the importance, especially of the liver and kidneys, in the exchange of Se in fish.
Expression of selected trout selenoprotein transcripts showed an inverse correlation with an increase in Sel-Plex in most cases. In liver, kidney, and blood cells, the highest activation of trout selenoprotein genes was observed mainly in the group receiving a diet enriched with the lowest Sel-Plex concentration (0.5 g/kg) for 10 weeks.
The study was carried out in order to clarify the dosage of selenium, based on the assumption that fish, in particular salmon, may require higher Se levels than mammals and more than the dosage allowed by current legislation (0.5 mg/kg dry feed weight).
A study by Mechlaoui et al. [56] was aimed at determining the effect of dietary inclusion of selenium (Se) in the form of inorganic Se (sodium selenite, Na2SeО3) and organic Se (hydroxyselenomethionine, HO-SeMet) on sea bream (gilthead seabream) (Sparus aurata). The control diet for fish (without Se supplementation) contained 0.8 mg selenium/kg feed and was used as a basal diet. Up to 2 groups of experimental feeds introduced selenium in the form of Na2SeО3 in an amount of 0.2 mg Se/kg and 0.5 mg Se/kg. Two more feed groups contained a similar amount of selenium in the form of OH-SeMet.
The highest growth rate was observed in fish fed OHSeMet at a level of 0.2 mg/kg, but without significant differences with fish which were given a control diet without adding Se. The smallest growth was observed in fish treated with sodium selenite, up to 0.5 mg/kg. An increase in Se in the diet, especially in the form of OH-SeMet, led to an increase in Se in the liver and muscles. The inclusion of OH-SeMet in the diet led to a significant (p<0.05) decrease in the content of malondialdehyde (MDA) in the liver and muscles. The inclusion of Se in the form of selenite at a dose of 0.2 mg/kg is not as effective as the organic supplement Se to improve the oxidative status of muscles. The dietary inclusion of Se at a dose of 0.2 mg/kg significantly reduced plasma cortisol levels after 2 hours of acute stress, regardless of what form of Se was given. Serum lysozyme activity decreased with an increase in the amount of added Se additives in the diet. Thus, the addition of Se to 0.2 mg/kg (1-1.1 mg/kg of the analyzed dietary Se), especially in the form of OHSeMet, had a beneficial effect on growth, maintaining liver morphology and improving fry protection (of juvenile) Dorado from acute or chronic stress. In addition, it was found that OH-SeMet is more effective than Na2SeO3 in protecting against oxidative stress in fish muscles.
The importance of introducing selenium supplements based on a plant ingredient for parent forms of rainbow trout had been demonstrated, which not only affected the reproductive functions of producers, but also ensured parental transmission of Se to offspring for the possibility of antioxidant metabolism from the very beginning of feeding [57]. Three groups of rainbow trout received a diet with or without selenium supplements (control, basal level of Se- 0.3 mg/kg of feed), a group with the addition of Se in the form of sodium selenite (Na2SeО3) in the amount of 0.3 mg/ kg of feed, as well as a third the group received food, in which selenium was additionally added in the form of OH-SeMet in an amount of 0.7 mg/kg of food. Trout was fed with this food for 6 months before spawning. In Se supplemented groups, the total number of spawning females was significantly higher compared to the negative control group, and females treated with OH-SeMet started spawning earlier than females treated with Na2SeО3 or control diet. Concentrations of total Se were significantly higher in the muscles of females from the group to which additional OH-SeMet was added. Higher concentrations of Se in oocytes of both groups with the addition of Se confirmed maternal transfer of Se, while the total concentration of Se in samples of seminal fluid did not differ significantly between the groups of fish. No effect was found on the activity of glutathione peroxidase (GPx) and other antioxidant enzymes in the offspring liver, while in hatched fry, GPX activity was significantly higher in both treatments supplemented with Se with the highest activity in the OH-SeMet group.
The Sе supplement enhances the expression of the hepatic SelPa gene of the uterine population of males and females along with the genes of such important selenoproteins as cytosolic and mitochondrial methionine sulfoxide reductase (MsrB1 and MsrB2), which play a critical role in protein redox regulation; glutathione peroxidase isoform genes (GPx1a, GPx4a2), catalase antioxidant enzyme (CAT), Glutamate cysteine ligase catalytic subunit (Gclc), glutamate cysteine ligase, also known as gamma glutamylcysteine synthetase K1, which is the first ECH associated protein 1), an element of the Keap1- Nrf2 signaling pathway, which is the main regulator of cytoprotective reactions to oxidative and electrophilic stress triggered by protein genes (MsrB1, GPx1a, GPx4a2, CAT, Gclc, Keap1) expressed in the liver of males.
In fry, which switched to active swimming, the addition of organic Se led to higher gene expression for SelPa, GPX1a, GPX1b2, CAT, and MsrB2. The improvement in oxidative status in the whole organism of fry was not confirmed in this study, since the ratio of reduced and oxidized glutathione (GSH/GSSG) was even lower in juvenile fish in groups supplemented with Se, while the levels of 8-isoprostane are a marker of oxidative stress activity were lower in the Na2SeO3 group. In addition, no effect was found on the levels of protein associated carbonyls, which are a marker of global protein oxidation, since they are generated by many different active forms of oxygen in the blood, tissues and cells.
Organic supplementation of Se led to a significant increase in the level of α-tocopherol and vitamin C in the offspring. These results show that the addition of selenium to broodstock feeds affects the stimulation and course of spawning, and the transfer of selenium from parents to offspring affects some features of the new generation.
The study of D. Pacitti et al. determined the expression of the constitutive mRNA of glutathione peroxidase (GPx) genes in different trout tissues and their reactions. Glutathione peroxidases (GPxs) are the largest and most studied family of selenoproteins. Cytosolic glutathione peroxidase (cGPx, GPx1) and phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) are widely distributed throughout the tissues and play a key role in regulating the oxidative status in the cell. The authors cloned the GPx1 and GPx4 genes in rainbow trout (Oncorhynchus mykiss). Accessibility to Se was analyzed using rainbow trout liver cell line (RTL). The non-organic form of selenium (sodium selenite Na2SeO3) and the organic form (selenocysteine, Cys-Se-Se-Cys) were used as Se sources. Activity was studied to test the effect of transcript changes on the enzymatic function of these molecules. To understand whether the results obtained from the analysis of transcript expression were due to the bioavailability of Se or the formation of reactive oxygen species (ROS), the cytotoxicity of the two selenium compounds was tested by measuring the effect of Se on the integrity of cell membranes. In addition, the bioavailability of Se was quantified by mass spectrometry to determine the amount of Se in cell culture media, and the contribution of the two selenium compounds used in the treatment. Three gene isoforms were identified for GPx1 (GPx1a, 1b1 and 1b2) and GPx4 (GPx4a1, a2 and b). The discovery of a third gene encoding GPx1 and GPx4 indicates that salmonids may have the largest selenoproteome among all vertebrates.
GPx4 gene transcripts were more pronounced in most tissues studied in vivo (except blood, head kidney, and spleen), while GPx1 genes were more sensitive to in vitro effects of selenium, especially to the organic form. Interestingly, GPx1a was most sensitive to selenium availability under non-stressful conditions, while GPx1b1 and GPx1b2 were strongly induced by exposure to selenium levels, which had some toxic effect on the cells. Although different test concentrations of the two selenium compounds modulate the expression of the GPx1 transcript, no significant changes in the enzymatic activity of GPx1 were detected to varying degrees. The results of these studies indicate that the expression level of transcripts of trout GPx1 may be a sensitive biomarker for selenium consumption, helping to assess whether selenium concentration and chemical speciation affect cell homeostasis.
The important role of selenium in increasing immunity in fish growth has been demonstrated. A growth study was conducted to determine the need for selenium in feed for black sea bream fish (Acanthopagrus schlegelii) in juvenile age [58]. The basal diet was supplemented with Se polysaccharide at levels of 0.34 mg/kg; 0.52; 0.68; 0.91; 1.08 and 3.06 mg/kg. Concentrations of Se ≤ 0.91 mg/kg of feed significantly influenced the increase in weight gain in fish, while higher levels of selenium incorporation showed a decrease in growth trends. The activity of superoxide dismutase, glutathione peroxidase, catalase, and glutathione reductase in serum and liver increased significantly and leveled out in fish fed ≥ 1.08 mg Se/kg ration, while the content of malondialdehyde in the liver and serum decreased significantly with increasing Se levels. The content of Se in the liver and muscles increased linearly with increasing levels of Se in the diet. Based on the results, it can be concluded that the need for Se diet for juvenile black sea bream is 0.86 mg/kg per weight gain.
Studies by Lee et al. [59] postulated the importance of the addition of trace elements, selenium (Se) to fish feed, because Se from the surrounding water and the feed itself cannot provide the optimal level required for cultivated aquatic species. Despite the fact that Se is usually required in much smaller quantities in the fish diet, but its addition at the optimum level is a critical problem in the formulation of fish feed. Literature data over the past two decades indicate that Se is sensitive enough and care should be taken to ensure that it is included at the optimum level in fish feed formulations. The experiment showed that juvenile Nile tilapia has special requirements for the presence of Se in the body, which cannot be provided with normal food or water from the environment. The inclusion of dietary Se at optimal concentrations is a prerequisite for effective growth, saturation of tissues and ensuring normal enzyme in Nile tilapia. The addition of Se to fish feed in excess of the required level can have toxic effects on freshwater aquaculture organisms.
Discussion
Researchers Wang et al. [60] proposed an experimental test of the hypothesis about the possibility of other selenoproteins, in addition to deiodinase, also participate in the regulation of fish growth. Selenium is an important trace element for fish growth and performs its physiological functions mainly by incorporation into selenoproteins. It is well known that dietary Se regulates fish growth by controlling the synthesis of deiodinase, a species of selenoproteins. Recently, bony fish have been described 41 selenoprotein. In this study, rainbow trout (Oncorhynchus mykiss) was fed with graduated Se levels (2, 4, or 6 mg/kg, from selenium yeast, Seyeast) for 10 weeks. At the end of the feeding test, fish growth and the expression of 28 selenoprotein genes in tissues were evaluated. The results showed that dietary Se-yeast significantly increased fish growth (P<0.05). Correlation analysis showed that the growth of rainbow trout significantly and positively correlated with only 4 genes of selenoprotein in the liver, but with all 11 differentially expressed genes of selenoprotein in muscles (P<0.05) does not cause oxidative stress in fish tissues. In addition, Se dietary supplements elicited an overall upregulation of selenoprotein gene expression in the liver (10 genes) or muscle (11 genes) (P<0.05), and it showed the strongest correlation with mRNA levels of the W-like selenoprotein gene in muscle (P<0.05). Selenoprotein W plays the role of glutathione (GSH) dependent antioxidant, which may be involved in the redox process.
These results indicate that nutritional supplements with Se yeast are useful for the growth of rainbow trout, and improved growth rates are closely related to the expression of muscle genes of selenoprotein, in particular, the selenoprotein W-like gene. This study reveals the importance of muscle gene expression of selenoproteins and offers a new concept for the regulatory mechanism of Se diet for fish growth.
The effect of bioaccumulation of selenium on tilapia of Mozambique was studied. Se induces an oxidative stress effect such as (of lipid peroxidation (LPO) and protein carbonyl (PCO) lipid peroxidation (LPO) and oxide modification of proteins and (PCO) in fish gills and liver exposed to Se. Se exposure increases the activity of SOD, GPx, GST, metallothionein, GSH and inhibits CAT activity [61].
When the organic selenium is introduced into the diet of barramundi (juvenile barramundi (Lates calcarifer), their weight, growth rate (final weight, specific growth rate and weight gain), as well as protein digestibility coefficient increases and the activity of glutathione peroxidase, creatinine kinase [62].
Conclusion
Thus, a review of the literature shows that Se interacts with the functional genome through the complex response structure of the metabolic network. The results suggest that adequate analysis and data management of integrated ohmic methods for models with controlled exposure to Se will help to develop a strategy for its effective application and to calculate the possible risks from exposure to Se, and in a broader sense serve as a working paradigm for experimental studies.
References
- Sies H, Berndt C, Jones, DP. Oxidative stress. Annu Rev Biochem 2017;86:715-748.
- Acharya A, Das I, Chandhok D, Saha T. Redox regulation in cancer: A double-edged sword with therapeutic potential. Oxid Med. Cell Longev 2010;3:23-34.
- Tsekhmistrenko O, Tsekhmistrenko S, Bityutskyy V. Nanoscale cerium dioxide as a mimetic of antioxidant protection enzymes. Multidisciplinary conference for young researchers 2019.
- Jones DP. Redefining oxidative stress. Antioxid Redox Signal 2008;8:1865-1879.
- Jones DP. Redox theory of aging. Redox. Biol 2015;5:71-79.
- Chris UO, Singh NB, Agarwal A. Nanoparticles as feed supplement on Growth behaviour of Cultured Catfish (Clarias gariepinus) fingerlings. Materials Today: Proceedings 2018;5:9076-9081.
- Ingold I, Berndt C, Schmitt S, Doll S, Poschmann G, Buday K, et al. Selenium utilization by GPX4 is required to prevent hydroperoxide-induced ferroptosis. Cell 2018;172:409-422.
- Bell JG, Cowey CB. Digestibility and bioavailability of dietary selenium from fishmeal, selenite, selenomethionine and selenocystine in Atlantic salmon (Salmo salar). Aquaculture 1989;81:61-68.
- Berntssen MH, Betancor M, Caballero MJ, Hillestad M, Rasinger J, Hamre K, et al. Safe limits of selenomethionine and selenite supplementation to plant-based Atlantic salmon feeds. Aquaculture 2018;495:617-630.
- Iswarya A, Vaseeharan B, Anjugam M, Gobi N, Divya M, Faggio C. Ã?-1, 3 glucan binding protein based selenium nanowire enhances the immune status of Cyprinus carpio and protection against Aeromonas hydrophila infection. Fish & shellfish immunology 2018;83:61â??75.
- Tsekhmistrenko SI, Bityutskyy VS, Tsekhmistrenko OS, Polishchuk VM, Polishchuk SA, Ponomarenko NV, et al. Enzyme-like activity of nanomaterials. Regul Mech Biosyst 2018;9:469-476.
- Brown DI, Griendling Kk. Regulation of signal transduction by reactive oxygen species in the cardiovascular system. Circulation research 2015;116:531-549.
- Misra BB, Langefeld CD, Olivier M, Cox LA. Integrated omics: tools, advances, and future approaches. J mol endocrinol 2018;62:21-45.
- Fernandes J, Hu X, Smith MR. Go YM, Jones DP. Selenium at the redox interface of the genome, metabolome and exposome. Free Radic Biol Med 2018;127:215-227.
- Speckmann B, Schulz S, Hiller F, Hesse D, Schumacher F, Kleuser B, et al. Selenium increases hepatic DNA methylation and modulates one-carbon metabolism in the liver of mice. J Nutr Biochem 2017;48:112-119.
- Maehira F, Miyagi I, Eguchi Y. Selenium regulates transcription factor NF-kappaB activation during the acute phase reaction. Clinica Chimica Acta 2003;334:163-171.
- Youn HS, Lim HJ, Choi YJ, Lee JY, Lee MY, Ryu JH. Selenium suppresses the activation of transcription factor NF-kappa B and IRF3 induced by TLR3 or TLR4 agonists. Int Immunopharmacol 2008 ;8:495-501.
- Ross AC. Modern nutrition in health and disease. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins 2014.
- Berry MJ, Banu L, Harney JW, Larsen PR. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. The EMBO Journal 1993;12:3315-3322.
- Lobanov AV, Hatfield DL, Gladyshev VN. Eukaryotic selenoproteins and selenoproteomes. Biochim Biophys Acta 2009;1790:1424-1428.
- Pacitti D, Wang T, Martin SAM, Sweetman J, Secombes CJ. Insights into the fish thioredoxin system: Expression profile of thioredoxin and thioredoxin reductase in rainbow trout (Oncorhynchus mykiss) during infection and in vitro stimulation. Dev Comp Immunol 2014;42:261-277.
- Bityutskyy VS, Tsekhmistrenko ?S, Tsekhmistrenko SI, Spyvack MY, Shadura UM. Perspectives of cerium nanoparticles use in agriculture. Animal Biology 2017;19:9-17.
- Betancor MB, Dam TM, Walton J, Morken T, Campbell PJ, Tocher DR. Modulation of selenium tissue distribution and selenoprotein expression in Atlantic salmon (Salmo salar L.) fed diets with graded levels of plant ingredients. British J Nutrition 2016;15:1325-1338.
- Ojeda ML, Carreras O, DÃaz Castro J, Murillo ML, Nogales F. High-and low-selenium diets affect endocrine energy balance during early programming. Toxicology and applied pharmacology 2019;382:114744.
- Ojeda ML, Nogales F, Membrilla A, Carreras O. Maternal selenium status is profoundly involved in metabolic fetal programming by modulating insulin resistance, oxidative balance and energy homeostasis. Euro J Nutri 2019;8:3171-3181.
- Brigelius Flohe R, Flohé L. Selenium and redox signaling. Archives of biochemistry and biophysics 2017;617:48-59.
- Cao N, Li W, Li B, Tian Y, Xu D. Transcriptome profiling reveals the immune response of goose T cells under selenium stimuli. Animal Sci J 2017;88:2001-2009.
- Cotter PA, Craig SR, Mclean E. Hyperaccumulation of selenium in hybrid striped bass: a functional food for aquaculture? Aquaculture Nutrition 2008;14:15-222.
- Pacitti D, Lawan MM, Sweetman J, Martin SA, Feldmann J, Secombes CJ. Selenium supplementation in fish: A combined chemical and biomolecular study to understand Sel-Plex assimilation and impact on selenoproteome expression in rainbow trout (Oncorhynchus mykiss). PLoS One 2015;10:e0127041.
- Mariotti M, Ridge PG, Zhang Y, Lobanov AV, Pringle TH, Guigo R, et al. Composition and evolution of the vertebrate and mammalian selenoproteomes. PloS one 2012;7:e33066.
- Pacitti D, Wang T, Page MM, Martin SAM, Sweetman J, Feldmann J, et al. Characterization of cytosolic glutathione peroxidase and phospholipid-hydroperoxide glutathione peroxidase genes in rainbow trout (Oncorhynchus mykiss) and their modulation by in vitro selenium exposure. Aquat Toxicol 2013;130:97-111.
- Malandrakis EE, Exadactylos A, Dadali O, Golomazou E, Klaoudatos S, Panagiotaki P. Molecular cloning of four glutathione peroxidase (GPx) homologs and expression analysis during stress exposure of the marine teleost Sparus aurata. Comp Biochem Physiol B Biochem Mol Biol 2014;168:53-61.
- Goldson AJ, Fairweather Tait SJ, Armah CN, Bao Y, Broadley MR, Dainty JR, et al. Effects of selenium supplementation on selenoprotein gene expression and response to influenza vaccine challenge: a randomised controlled trial. PLoS One 2011;6:e14771.
- Hatfield DL, Tsuji PA, Carlson BA, Gladyshev VN. Selenium and selenocysteine: Roles in cancer, health, and development. Trends in biochemical sciences 2014;39:112-120.
- Jovanovic A, Grubor Lajsic G, Djukic N, Gardinovacki G, Matic A, Spasic M. The effect of selenium on antioxidant system in erythrocytes and liver of the carp (Cyprinus carpio L.). Crit Rev Food Sci Nutr 1997;37:443-448.
- Köhrle J, Schweizer U, Schomburg L. Selenium transport in mammals: Selenoprotein P and its receptors. Selenium 2012;205-219.
- Zavacki AM, Marsili A, Larsen PR. Control of thyroid hormone activation and inactivation by the iodothyronine deiodinase family of selenoenzymes. Selenium 2012;369â??381.
- Surai PF, Kochish II. Nutritional modulation of the antioxidant capacities in poultry: The case of selenium. Poultry science 2019;98:4231-4239.
- Gangadoo S, Stanley D, Hughes RJ, Moore RJ, Chapman J. The synthesis and characterisation of highly stable and reproducible selenium nanoparticles. Inorganic and Nano-Metal Chemistry 2017;47:1568â??1576.
- Tymoshok NO, Kharchuk MS, Kaplunenko VG, Bityutskyy VS, Tsekhmistrenko SI, Tsekhmistrenk, et al. Evaluation of effects of selenium nanoparticles on Bacillus subtilis. Regul Mech Biosyst 2019;10:544-552.
- Bityutskyy V, Tsekhmistrenko S, Tsekhmistrenko O, Melnychenko O, Kharchyshyn V. Effects of different dietary selenium sources including probiotics mixture on growth performance, feed utilization and serum biochemical profile of quails. In: Nadykto V. (eds) Modern Development Paths of Agricultural Production. Springer, Cham 2019;623-632.
- Tsekhmistrenko ?, Bityutskyy V, Tsekhmistrenko S, Melnychenko O, Tymoshok N, Spivak M. Use of nanoparticles of metals and non-metals in poultry farming. Animal Husbandry Products Production and Processing 2019;2:113-130.
- Ahmed SMU, Luo L, Namani A, Wang XJ, Tang X. Nrf2 signaling pathway: Pivotal roles in inflammation. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 2017;1863:585-597.
- Ashouriz S, Keyvanshokooh S, Salati AP, Johari SA, Pasha-Zanoosi H. Effects of different levels of dietary selenium nanoparticles on growth performance, muscle composition, blood biochemical profiles and antioxidant status of common carp (Cyprinus carpio). Aquaculture 2015;446:25-29.
- Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, et al. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3'-untranslated region. Nature 1991;353:273-276.
- Chambers I, Frampton J, Goldfarb P, Affara N, McBain W, Harrison PR. The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the â??terminationâ?? codon, TGA. EMBO J 1986;5:1221-1227.
- Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J 2000;19:306-314.
- Forchhammer K, Leinfelder W, Bock A. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature 1989;342:453-456.
- Maroney MJ, Hondal RJ. Selenium versus sulfur: Reversibility of chemical reactions and resistance to permanent oxidation in proteins and nucleic acids. Free Radic Biol Med 2018;127:228-237.
- Adams AL, Macmillan D. Investigation of peptide thioester formation via N?Se acyl transfer. J Pept Sci. 2013;19:65-73.
- Hamilton SJ. Review of residue-based selenium toxicity thresholds for freshwater fish. Ecotoxicology and Environmental safety 2003;56:201-210.
- Scienti?c opinion on dietary reference values for selenium. EFSA J 2014;12:3846.
- Scienti?c Opinion on Safety and E?cacy of Selenium in the Form of Organic Compounds Produced by the Selenium-enriched Yeast Saccharomyces Cerevisiae NCYC R646 (Selemax 1000/2000) as Feed Additive for All Species. The European Food Safety Authority (EFSA) Panel on Additives and Products or Substances used in Animal Feed (FEEDAP), Parma, Italy, EFSA J 2012;10:2778.
- Sele V, Ã?rnsrud R, Sloth JJ, Berntssen MH, Amlund H. Selenium and selenium species in feeds and muscle tissue of Atlantic salmon. J Trace Elem Med Biol 2018;47:124-133
- Pacitti D, Lawan MM, Feldmann J, Sweetman J, Wang T, Martin SAM, et al. Impact of selenium supplementation on fish antiviral responses: a whole transcriptomic analysis in rainbow trout (Oncorhynchus mykiss) fed supranutritional levels of Sel-Plex®. BMC genomics 2016;17:116.
- Mechlaoui M, Dominguez D, Robaina L, Geraert PA, Kaushik S, Saleh R, et al. Effects of different dietary selenium sources on growth performance, liver and muscle composition, antioxidant status, stress response and expression of related genes in gilthead seabream (Sparus aurata). Aquaculture 2019;507:251-259.
- Wischhusen P, Parailloux M, Geraert PA, Briens M, Bueno M, Mounicou S, et al. Effect of dietary Se in rainbow trout (Oncorhynchus mykiss) broodstock on antioxidant status, its parental transfer and oxidative status in the progeny. Aquaculture 2019;507:126-138.
- Wang L, Xiao JX, Hua Y, Xiang XW, Zhou YF, Ye L, et al. Effects of dietary selenium polysaccharide on growth performance, oxidative stress and tissue selenium accumulation of juvenile black sea bream, Acanthopagrus schlegelii. Aquaculture 2019;503:389-395.
- Lee S, Nambi RW, Won S, Katya K, Bai SC. Dietary selenium requirement and toxicity levels in juvenile Nile tilapia, Oreochromis niloticus. Aquaculture 2016;464:153-158.
- Wang L, Zhang X, Wu L, Liu Q, Zhang D, Yin J. Expression of selenoprotein genes in muscle is crucial for the growth of rainbow trout (Oncorhynchus mykiss) fed diets supplemented with selenium yeast. Aquaculture 2018;492:82-90.
- Gobi N, Vaseeharan B, Rekha R, Vijayakumar S, Faggio C. Bioaccumulation, cytotoxicity and oxidative stress of the acute exposure selenium in Oreochromis mossambicus. Ecotoxicology and environmental safety 2018;162:147-159.
- Ilham I, Fotedar R. Growth, enzymatic glutathione peroxidase activity and biochemical status of juvenile barramundi (Lates calcarifer) fed dietary fermented soybean meal and organic selenium. Fish physiology and biochemistry 2017;43:775-790.